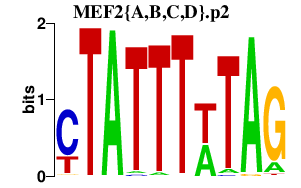
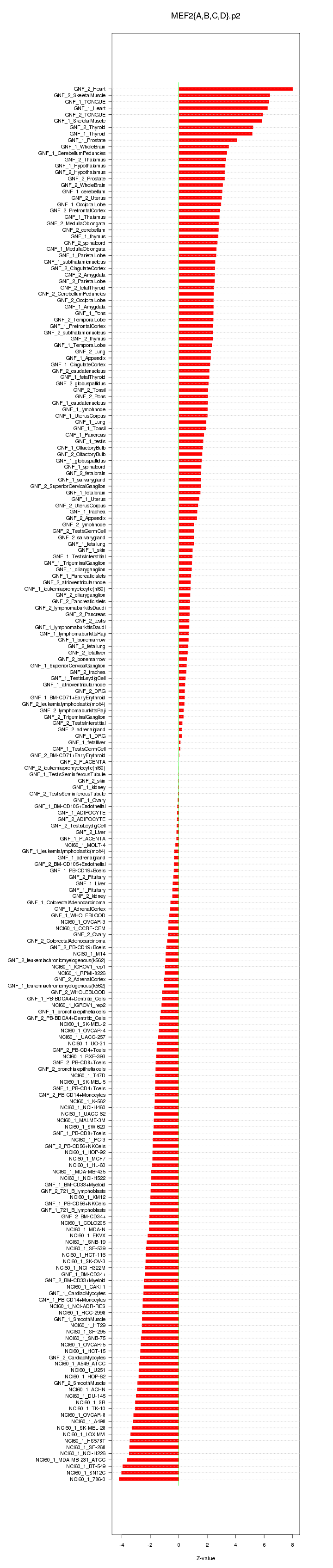
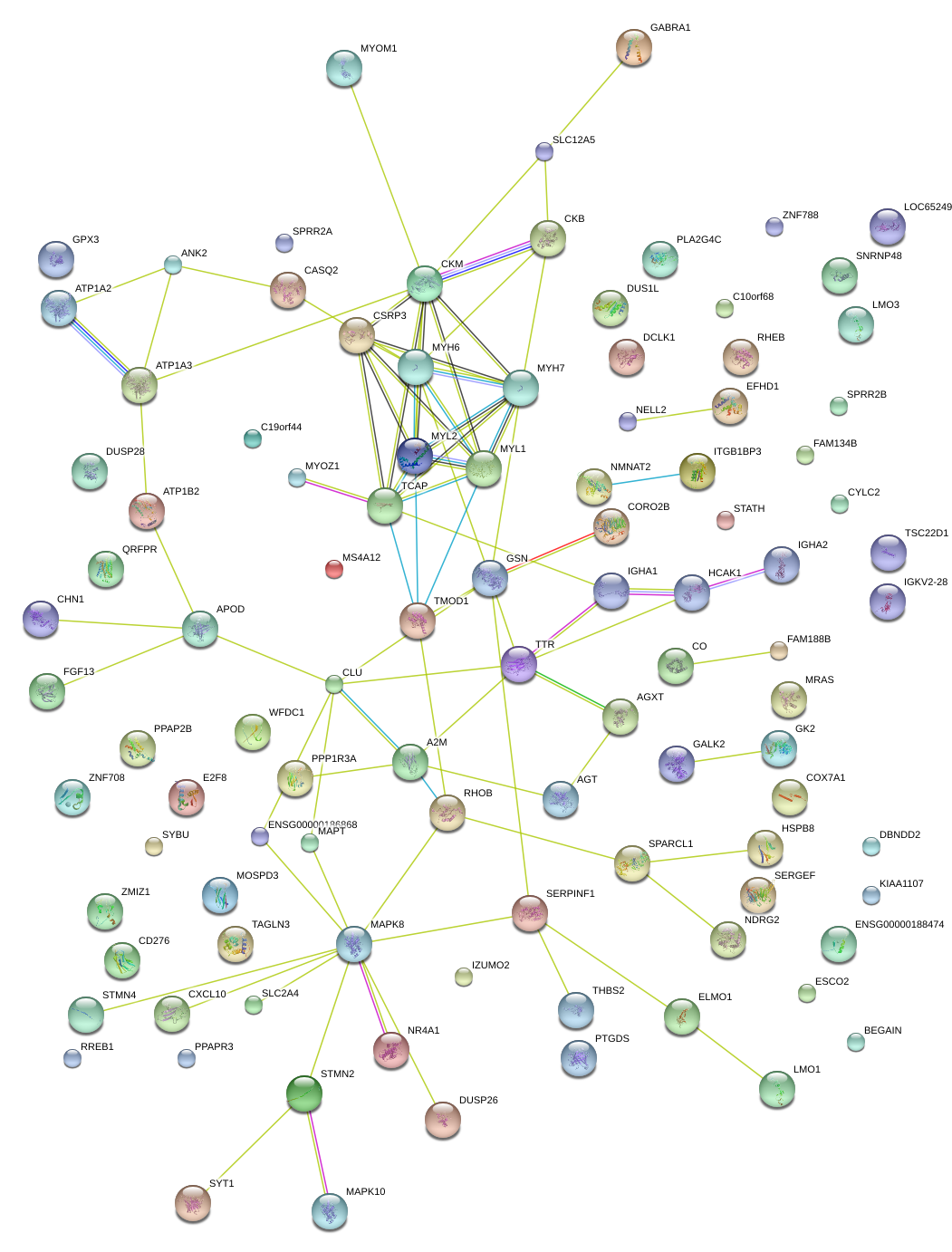
|
chr4_-_88450601
|
85.166
|
NM_001128310
NM_004684
|
SPARCL1
|
SPARC-like 1 (hevin)
|
|
chr9_+_139871954
|
52.106
|
NM_000954
|
PTGDS
|
prostaglandin D2 synthase 21kDa (brain)
|
|
chr5_-_42811959
|
48.647
|
NM_001085486
NM_001093726
NM_005410
|
SEPP1
|
selenoprotein P, plasma, 1
|
|
chr13_-_45048436
|
47.509
|
NM_001243798
|
TSC22D1
|
TSC22 domain family, member 1
|
|
chr9_+_139872014
|
47.143
|
|
PTGDS
|
prostaglandin D2 synthase 21kDa (brain)
|
|
chr12_+_52445185
|
45.517
|
NM_002135
NM_173157
|
NR4A1
|
nuclear receptor subfamily 4, group A, member 1
|
|
chr12_-_45269340
|
43.145
|
NM_001145109
|
NELL2
|
NEL-like 2 (chicken)
|
|
chr12_-_9268440
|
40.265
|
|
A2M
|
alpha-2-macroglobulin
|
|
chr12_-_16759939
|
39.524
|
NM_001243613
|
LMO3
|
LIM domain only 3 (rhombotin-like 2)
|
|
chr8_-_27472188
|
39.468
|
|
CLU
|
clusterin
|
|
chr14_-_21493858
|
38.581
|
|
NDRG2
|
NDRG family member 2
|
|
chr12_-_111358353
|
37.469
|
NM_000432
|
MYL2
|
myosin, light chain 2, regulatory, cardiac, slow
|
|
chr14_-_21493910
|
37.200
|
NM_016250
NM_201535
NM_201536
NM_201537
|
NDRG2
|
NDRG family member 2
|
|
chr17_+_37821598
|
35.602
|
NM_003673
|
TCAP
|
titin-cap (telethonin)
|
|
chr1_+_160085519
|
32.461
|
NM_000702
|
ATP1A2
|
ATPase, Na+/K+ transporting, alpha 2 polypeptide
|
|
chr20_+_44657812
|
31.925
|
NM_020708
|
SLC12A5
|
solute carrier family 12 (potassium/chloride transporter), member 5
|
|
chr2_-_211168315
|
31.864
|
NM_079422
|
MYL1
|
myosin, light chain 1, alkali; skeletal, fast
|
|
chr20_+_44657858
|
31.810
|
|
SLC12A5
|
solute carrier family 12 (potassium/chloride transporter), member 5
|
|
chr1_+_92632608
|
31.456
|
NM_015237
|
KIAA1107
|
KIAA1107
|
|
chr14_-_103989195
|
30.898
|
NM_001823
|
CKB
|
creatine kinase, brain
|
|
chr5_+_150399980
|
30.863
|
NM_002084
|
GPX3
|
glutathione peroxidase 3 (plasma)
|
|
chr2_+_233498206
|
29.922
|
NM_025202
|
EFHD1
|
EF-hand domain family, member D1
|
|
chr19_-_36643770
|
29.405
|
NM_001864
|
COX7A1
|
cytochrome c oxidase subunit VIIa polypeptide 1 (muscle)
|
|
chr8_-_33457491
|
28.687
|
|
DUSP26
|
dual specificity phosphatase 26 (putative)
|
|
chr14_-_103989112
|
28.425
|
|
CKB
|
creatine kinase, brain
|
|
chr6_+_7108864
|
28.087
|
|
RREB1
|
ras responsive element binding protein 1
|
|
chr18_-_3219986
|
27.955
|
NM_003803
NM_019856
|
MYOM1
|
myomesin 1, 185kDa
|
|
chr19_+_12203077
|
27.926
|
|
ZNF788
|
zinc finger family member 788
|
|
chr5_+_150400132
|
27.247
|
|
GPX3
|
glutathione peroxidase 3 (plasma)
|
|
chr1_-_230849863
|
27.180
|
|
AGT
|
angiotensinogen (serpin peptidase inhibitor, clade A, member 8)
|
|
chr1_-_116311161
|
26.580
|
|
CASQ2
|
calsequestrin 2 (cardiac muscle)
|
|
chr1_-_116311398
|
26.523
|
NM_001232
|
CASQ2
|
calsequestrin 2 (cardiac muscle)
|
|
chr1_-_116311330
|
25.689
|
|
CASQ2
|
calsequestrin 2 (cardiac muscle)
|
|
chr19_-_45822841
|
25.655
|
|
CKM
|
creatine kinase, muscle
|
|
chr16_+_84328400
|
25.645
|
NM_021197
|
WFDC1
|
WAP four-disulfide core domain 1
|
|
chr7_+_30951414
|
25.567
|
NM_198098
|
AQP1
|
aquaporin 1 (Colton blood group)
|
|
chr8_-_33457279
|
25.138
|
|
DUSP26
|
dual specificity phosphatase 26 (putative)
|
|
chr11_-_19223516
|
24.940
|
NM_001127656
NM_003476
|
CSRP3
|
cysteine and glycine-rich protein 3 (cardiac LIM protein)
|
|
chr12_-_9268475
|
24.907
|
|
A2M
|
alpha-2-macroglobulin
|
|
chr1_-_57045129
|
24.374
|
|
PPAP2B
|
phosphatidic acid phosphatase type 2B
|
|
chr14_-_103988829
|
24.130
|
|
CKB
|
creatine kinase, brain
|
|
chr12_-_16760935
|
24.128
|
NM_001001395
NM_001243609
|
LMO3
|
LIM domain only 3 (rhombotin-like 2)
|
|
chr2_+_20646831
|
24.096
|
NM_004040
|
RHOB
|
ras homolog gene family, member B
|
|
chr12_+_119616594
|
23.704
|
NM_014365
|
HSPB8
|
heat shock 22kDa protein 8
|
|
chr5_+_161274939
|
23.559
|
NM_001127644
|
GABRA1
|
gamma-aminobutyric acid (GABA) A receptor, alpha 1
|
|
chr8_-_27115935
|
23.542
|
|
STMN4
|
stathmin-like 4
|
|
chr20_+_44035203
|
22.762
|
NM_001048223
NM_001048224
|
DBNDD2
|
dysbindin (dystrobrevin binding protein 1) domain containing 2
|
|
chr13_-_45011389
|
22.579
|
NM_001243797
|
TSC22D1
|
TSC22 domain family, member 1
|
|
chr17_+_7554711
|
21.979
|
|
ATP1B2
|
ATPase, Na+/K+ transporting, beta 2 polypeptide
|
|
chr7_-_37488554
|
21.866
|
NM_014800
|
ELMO1
|
engulfment and cell motility 1
|
|
chr8_+_27631902
|
21.801
|
NM_001017420
|
ESCO2
|
establishment of cohesion 1 homolog 2 (S. cerevisiae)
|
|
chr8_-_110661007
|
21.754
|
NM_001099747
NM_001099748
NM_001099749
NM_001099750
NM_017786
|
SYBU
|
syntabulin (syntaxin-interacting)
|
|
chr4_-_88450243
|
21.680
|
|
SPARCL1
|
SPARC-like 1 (hevin)
|
|
chr10_-_75401199
|
21.359
|
|
MYOZ1
|
myozenin 1
|
|
chr12_-_16758312
|
21.355
|
NM_001243611
|
LMO3
|
LIM domain only 3 (rhombotin-like 2)
|
|
chr7_-_64023444
|
21.185
|
|
|
|
|
chr6_-_169653922
|
21.001
|
|
THBS2
|
thrombospondin 2
|
|
chr22_+_22569155
|
20.918
|
|
|
|
|
chr13_-_45011059
|
20.555
|
NM_006022
|
TSC22D1
|
TSC22 domain family, member 1
|
|
chr3_-_195310770
|
20.553
|
|
APOD
|
apolipoprotein D
|
|
chr15_+_68908878
|
20.435
|
NM_001190456
|
CORO2B
|
coronin, actin binding protein, 2B
|
|
chr14_-_101036130
|
20.404
|
NM_020836
|
BEGAIN
|
brain-enriched guanylate kinase-associated homolog (rat)
|
|
chr10_-_75401492
|
20.286
|
NM_021245
|
MYOZ1
|
myozenin 1
|
|
chr17_+_43971655
|
20.026
|
NM_001123066
NM_001123067
NM_001203251
NM_001203252
NM_005910
NM_016834
NM_016835
NM_016841
|
MAPT
|
microtubule-associated protein tau
|
|
chr8_+_80523304
|
19.952
|
|
STMN2
|
stathmin-like 2
|
|
chr7_-_37488408
|
19.776
|
|
ELMO1
|
engulfment and cell motility 1
|
|
chr5_+_161275185
|
19.756
|
|
GABRA1
|
gamma-aminobutyric acid (GABA) A receptor, alpha 1
|
|
chr8_-_27115897
|
19.647
|
NM_030795
|
STMN4
|
stathmin-like 4
|
|
chr18_-_3219846
|
19.598
|
|
MYOM1
|
myomesin 1, 185kDa
|
|
chr11_+_60260250
|
19.283
|
NM_001164470
NM_017716
|
MS4A12
|
membrane-spanning 4-domains, subfamily A, member 12
|
|
chr4_-_122302112
|
19.063
|
NM_198179
|
QRFPR
|
pyroglutamylated RFamide peptide receptor
|
|
chr17_+_1665306
|
18.956
|
|
SERPINF1
|
serpin peptidase inhibitor, clade F (alpha-2 antiplasmin, pigment epithelium derived factor), member 1
|
|
chr3_+_111717585
|
18.466
|
NM_001008272
NM_013259
|
TAGLN3
|
transgelin 3
|
|
chr14_-_107131217
|
18.314
|
|
IGHA1
|
immunoglobulin heavy constant alpha 1
|
|
chrX_-_138287180
|
18.116
|
NM_001139498
NM_001139500
NM_001139501
|
FGF13
|
fibroblast growth factor 13
|
|
chr1_-_183274008
|
17.454
|
NM_170706
|
NMNAT2
|
nicotinamide nucleotide adenylyltransferase 2
|
|
chr9_+_124048396
|
17.300
|
|
GSN
|
gelsolin
|
|
chr17_+_1665342
|
17.207
|
|
SERPINF1
|
serpin peptidase inhibitor, clade F (alpha-2 antiplasmin, pigment epithelium derived factor), member 1
|
|
chr4_+_113970784
|
17.160
|
NM_001148
NM_020977
|
ANK2
|
ankyrin 2, neuronal
|
|
chr19_+_16607181
|
17.113
|
NM_032207
|
C19orf44
|
chromosome 19 open reading frame 44
|
|
chr12_-_9268513
|
16.898
|
|
A2M
|
alpha-2-macroglobulin
|
|
chr17_+_1665281
|
16.815
|
|
SERPINF1
|
serpin peptidase inhibitor, clade F (alpha-2 antiplasmin, pigment epithelium derived factor), member 1
|
|
chr1_-_57045235
|
16.656
|
NM_003713
|
PPAP2B
|
phosphatidic acid phosphatase type 2B
|
|
chr8_+_80523351
|
16.582
|
|
STMN2
|
stathmin-like 2
|
|
chr17_+_1665233
|
16.564
|
NM_002615
|
SERPINF1
|
serpin peptidase inhibitor, clade F (alpha-2 antiplasmin, pigment epithelium derived factor), member 1
|
|
chr2_-_175712276
|
16.554
|
NM_001206602
|
CHN1
|
chimerin (chimaerin) 1
|
|
chr5_-_16509106
|
16.501
|
NM_019000
|
FAM134B
|
family with sequence similarity 134, member B
|
|
chr19_-_50666279
|
16.433
|
NM_152358
|
IZUMO2
|
IZUMO family member 2
|
|
chr4_-_76944585
|
16.424
|
NM_001565
|
CXCL10
|
chemokine (C-X-C motif) ligand 10
|
|
chr7_-_151216951
|
16.282
|
NM_005614
|
RHEB
|
Ras homolog enriched in brain
|
|
chr7_-_113559048
|
16.100
|
NM_002711
|
PPP1R3A
|
protein phosphatase 1, regulatory subunit 3A
|
|
chr18_+_29171729
|
15.925
|
NM_000371
|
TTR
|
transthyretin
|
|
chr2_-_89292396
|
15.770
|
|
IGKC
|
immunoglobulin kappa constant
|
|
chr2_-_211179765
|
15.719
|
NM_079420
|
MYL1
|
myosin, light chain 1, alkali; skeletal, fast
|
|
chr9_+_103947316
|
15.657
|
NM_017753
|
LPPR1
|
lipid phosphate phosphatase-related protein type 1
|
|
chr1_-_230849847
|
15.386
|
|
AGT
|
angiotensinogen (serpin peptidase inhibitor, clade A, member 8)
|
|
chr19_+_3933100
|
15.340
|
NM_170678
|
ITGB1BP3
|
integrin beta 1 binding protein 3
|
|
chr3_+_138067678
|
15.279
|
NM_001252092
|
MRAS
|
muscle RAS oncogene homolog
|
|
chr15_+_73976720
|
15.167
|
|
CD276
|
CD276 molecule
|
|
chr9_+_100263818
|
15.125
|
NM_003275
|
TMOD1
|
tropomodulin 1
|
|
chr1_-_230850335
|
15.003
|
NM_000029
|
AGT
|
angiotensinogen (serpin peptidase inhibitor, clade A, member 8)
|
|
chr3_+_138067444
|
14.983
|
NM_001085049
NM_001252093
|
MRAS
|
muscle RAS oncogene homolog
|
|
chr12_-_9268522
|
14.956
|
NM_000014
|
A2M
|
alpha-2-macroglobulin
|
|
chr12_-_45270157
|
14.776
|
NM_001145108
NM_006159
|
NELL2
|
NEL-like 2 (chicken)
|
|
chr1_-_57044980
|
14.758
|
|
PPAP2B
|
phosphatidic acid phosphatase type 2B
|
|
chr4_-_87028805
|
14.640
|
NM_138981
|
MAPK10
|
mitogen-activated protein kinase 10
|
|
chr1_-_153029987
|
14.600
|
NM_005988
|
SPRR2A
|
small proline-rich protein 2A
|
|
chr19_-_21512148
|
14.544
|
NM_021269
|
ZNF708
|
zinc finger protein 708
|
|
chr8_-_27472298
|
14.499
|
NM_001831
|
CLU
|
clusterin
|
|
chr12_-_16759531
|
14.426
|
NM_018640
|
LMO3
|
LIM domain only 3 (rhombotin-like 2)
|
|
chr10_+_80899458
|
14.333
|
|
ZMIZ1
|
zinc finger, MIZ-type containing 1
|
|
chr1_+_15990300
|
14.297
|
|
|
|
|
chr4_+_70861647
|
14.290
|
NM_001009181
NM_003154
|
STATH
|
statherin
|
|
chr12_+_79257772
|
14.288
|
NM_001135805
|
SYT1
|
synaptotagmin I
|
|
chr4_-_80329354
|
14.285
|
NM_033214
|
GK2
|
glycerol kinase 2
|
|
chr17_+_7184978
|
14.222
|
NM_001042
|
SLC2A4
|
solute carrier family 2 (facilitated glucose transporter), member 4
|
|
chr12_-_16759430
|
14.137
|
|
LMO3
|
LIM domain only 3 (rhombotin-like 2)
|
|
chr10_+_32856650
|
14.087
|
NM_024688
|
C10orf68
|
chromosome 10 open reading frame 68
|
|
chr6_+_7590405
|
13.991
|
NM_152551
|
SNRNP48
|
small nuclear ribonucleoprotein 48kDa (U11/U12)
|
|
chr14_-_23877480
|
13.915
|
NM_002471
|
MYH6
|
myosin, heavy chain 6, cardiac muscle, alpha
|
|
chr11_-_18034688
|
13.837
|
|
SERGEF
|
secretion regulating guanine nucleotide exchange factor
|
|
chr19_-_48614013
|
13.705
|
NM_001159322
NM_001159323
NM_003706
|
PLA2G4C
|
phospholipase A2, group IVC (cytosolic, calcium-independent)
|
|
chr13_-_36429798
|
13.689
|
NM_001195415
NM_001195416
NM_001195430
|
DCLK1
|
doublecortin-like kinase 1
|
|
chr9_+_105757592
|
13.664
|
NM_001340
|
CYLC2
|
cylicin, basic protein of sperm head cytoskeleton 2
|
|
chr7_+_23286390
|
13.536
|
|
GPNMB
|
glycoprotein (transmembrane) nmb
|
|
chr6_+_27806322
|
13.525
|
NM_003520
|
HIST1H2BN
|
histone cluster 1, H2bn
|
|
chr22_-_37215516
|
13.514
|
NM_002854
|
PVALB
|
parvalbumin
|
|
chr15_+_42651697
|
13.276
|
NM_000070
NM_024344
NM_173087
|
CAPN3
|
calpain 3, (p94)
|
|
chr1_+_13911966
|
13.220
|
NM_001006624
NM_001006625
|
PDPN
|
podoplanin
|
|
chr2_+_173686314
|
13.214
|
NM_001100397
|
RAPGEF4
|
Rap guanine nucleotide exchange factor (GEF) 4
|
|
chr7_-_56160604
|
13.190
|
NM_006213
|
PHKG1
|
phosphorylase kinase, gamma 1 (muscle)
|
|
chr6_-_27806116
|
13.173
|
NM_003510
|
HIST1H2AK
|
histone cluster 1, H2ak
|
|
chr10_+_88428263
|
13.105
|
NM_001080116
NM_001080114
NM_001080115
NM_001171611
NM_007078
|
LDB3
|
LIM domain binding 3
|
|
chr2_+_170366211
|
13.057
|
NM_006063
|
KBTBD10
|
kelch repeat and BTB (POZ) domain containing 10
|
|
chr4_-_39640461
|
13.051
|
NM_174921
|
C4orf34
|
chromosome 4 open reading frame 34
|
|
chr11_+_46958228
|
12.888
|
NM_001003676
NM_001003677
NM_001003678
NM_024113
|
C11orf49
|
chromosome 11 open reading frame 49
|
|
chr5_-_149535138
|
12.846
|
|
PDGFRB
|
platelet-derived growth factor receptor, beta polypeptide
|
|
chr16_-_31439650
|
12.750
|
NM_005205
|
COX6A2
|
cytochrome c oxidase subunit VIa polypeptide 2
|
|
chr5_-_115910406
|
12.736
|
NM_020796
|
SEMA6A
|
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6A
|
|
chr9_-_34691135
|
12.647
|
NM_006274
|
CCL19
|
chemokine (C-C motif) ligand 19
|
|
chr12_+_69201979
|
12.545
|
|
MDM2
|
Mdm2 p53 binding protein homolog (mouse)
|
|
chr11_-_128894008
|
12.433
|
NM_014715
|
ARHGAP32
|
Rho GTPase activating protein 32
|
|
chr11_-_47374221
|
12.431
|
NM_000256
|
MYBPC3
|
myosin binding protein C, cardiac
|
|
chr1_+_151032880
|
12.378
|
|
MLLT11
|
myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila); translocated to, 11
|
|
chr1_+_151032866
|
12.308
|
|
MLLT11
|
myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila); translocated to, 11
|
|
chr19_-_55668956
|
12.274
|
NM_000363
|
TNNI3
|
troponin I type 3 (cardiac)
|
|
chr12_-_16759733
|
12.254
|
NM_001243610
|
LMO3
|
LIM domain only 3 (rhombotin-like 2)
|
|
chr17_-_34891262
|
12.141
|
NM_001033580
NM_001163735
NM_025109
|
MYO19
|
myosin XIX
|
|
chr13_-_25497054
|
12.059
|
NM_018451
|
CENPJ
|
centromere protein J
|
|
chr22_-_38577692
|
12.044
|
NM_001004426
NM_003560
NM_001199562
|
PLA2G6
|
phospholipase A2, group VI (cytosolic, calcium-independent)
|
|
chr1_+_183605207
|
11.984
|
NM_015149
|
RGL1
|
ral guanine nucleotide dissociation stimulator-like 1
|
|
chr12_+_81101407
|
11.980
|
NM_002469
|
MYF6
|
myogenic factor 6 (herculin)
|
|
chr10_-_14879967
|
11.883
|
NM_001029954
|
CDNF
|
cerebral dopamine neurotrophic factor
|
|
chr11_-_18034491
|
11.839
|
NM_012139
|
SERGEF
|
secretion regulating guanine nucleotide exchange factor
|
|
chr19_-_20748540
|
11.732
|
NM_001159293
|
ZNF737
|
zinc finger protein 737
|
|
chr1_+_65730423
|
11.647
|
NM_014787
|
DNAJC6
|
DnaJ (Hsp40) homolog, subfamily C, member 6
|
|
chr10_-_45474210
|
11.605
|
NM_007021
|
C10orf10
|
chromosome 10 open reading frame 10
|
|
chr6_-_6007097
|
11.414
|
|
NRN1
|
neuritin 1
|
|
chr17_-_2966900
|
11.373
|
NM_014566
|
OR1D5
|
olfactory receptor, family 1, subfamily D, member 5
|
|
chr4_-_186877413
|
11.369
|
|
SORBS2
|
sorbin and SH3 domain containing 2
|
|
chrX_+_64708614
|
11.181
|
NM_001010888
|
ZC3H12B
|
zinc finger CCCH-type containing 12B
|
|
chr15_+_41576150
|
11.160
|
|
OIP5-AS1
|
OIP5 antisense RNA 1 (non-protein coding)
|
|
chr1_-_201390845
|
11.156
|
NM_003281
|
TNNI1
|
troponin I type 1 (skeletal, slow)
|
|
chr5_+_53751430
|
11.102
|
NM_006308
|
HSPB3
|
heat shock 27kDa protein 3
|
|
chr19_+_18062110
|
11.015
|
NM_002248
|
KCNN1
|
potassium intermediate/small conductance calcium-activated channel, subfamily N, member 1
|
|
chr9_+_21440452
|
10.989
|
NM_024013
|
IFNA1
|
interferon, alpha 1
|
|
chr9_-_21228132
|
10.970
|
NM_021268
|
IFNA17
|
interferon, alpha 17
|
|
chr15_+_93443418
|
10.837
|
NM_001042572
NM_001271
|
CHD2
|
chromodomain helicase DNA binding protein 2
|
|
chr7_-_99680358
|
10.820
|
|
ZNF3
|
zinc finger protein 3
|
|
chrX_-_21776158
|
10.767
|
NM_014332
|
SMPX
|
small muscle protein, X-linked
|
|
chr1_+_160160284
|
10.656
|
NM_001231
|
CASQ1
|
calsequestrin 1 (fast-twitch, skeletal muscle)
|
|
chr11_+_1860232
|
10.646
|
NM_003282
|
TNNI2
|
troponin I type 2 (skeletal, fast)
|
|
chr4_+_41362750
|
10.588
|
NM_001112717
NM_001112718
NM_014988
|
LIMCH1
|
LIM and calponin homology domains 1
|
|
chr5_+_140749830
|
10.587
|
NM_018924
NM_032097
|
PCDHGB3
|
protocadherin gamma subfamily B, 3
|
|
chr1_+_151032683
|
10.579
|
|
MLLT11
|
myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila); translocated to, 11
|
|
chr7_+_23286386
|
10.562
|
|
GPNMB
|
glycoprotein (transmembrane) nmb
|
|
chr4_-_46995440
|
10.505
|
NM_001204266
NM_001204267
|
GABRA4
|
gamma-aminobutyric acid (GABA) A receptor, alpha 4
|
|
chr14_+_95078713
|
10.501
|
NM_001085
|
SERPINA3
|
serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 3
|
|
chr8_-_6856678
|
10.478
|
NM_004084
NM_001042500
|
DEFA1
DEFA1B
|
defensin, alpha 1
defensin, alpha 1B
|
|
chr12_-_371994
|
10.476
|
NM_001190997
NM_001243392
NM_016615
|
SLC6A13
|
solute carrier family 6 (neurotransmitter transporter, GABA), member 13
|
|
chr12_-_45270071
|
10.449
|
|
NELL2
|
NEL-like 2 (chicken)
|
|
chr9_+_33290536
|
10.327
|
|
NFX1
|
nuclear transcription factor, X-box binding 1
|
|
chr3_-_114343052
|
10.302
|
NM_001164347
|
ZBTB20
|
zinc finger and BTB domain containing 20
|
|
chr2_+_33661392
|
10.202
|
NM_170672
|
RASGRP3
|
RAS guanyl releasing protein 3 (calcium and DAG-regulated)
|
|
chr7_-_37488894
|
10.196
|
NM_001206480
|
ELMO1
|
engulfment and cell motility 1
|
|
chr10_-_69597768
|
10.181
|
NM_021800
NM_201262
|
DNAJC12
|
DnaJ (Hsp40) homolog, subfamily C, member 12
|
|
chr4_-_140098236
|
10.175
|
|
|
|
|
chr10_+_88428167
|
10.070
|
NM_001171610
|
LDB3
|
LIM domain binding 3
|
|
chr1_-_115238091
|
10.054
|
NM_000036
NM_001172626
|
AMPD1
|
adenosine monophosphate deaminase 1
|
|
chr5_-_142077508
|
10.011
|
NM_001144934
NM_001144935
|
FGF1
|
fibroblast growth factor 1 (acidic)
|
|
chr3_+_9691184
|
9.920
|
|
MTMR14
|
myotubularin related protein 14
|
|
chr19_-_54311958
|
9.877
|
NM_033297
|
NLRP12
|
NLR family, pyrin domain containing 12
|
|
chr10_-_75415748
|
9.860
|
NM_001114133
|
SYNPO2L
|
synaptopodin 2-like
|
|
chr5_+_140729723
|
9.854
|
NM_018922
NM_032095
|
PCDHGB1
|
protocadherin gamma subfamily B, 1
|
|
chr7_-_83278478
|
9.752
|
NM_012431
|
SEMA3E
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3E
|
|
chr1_-_153085963
|
9.738
|
NM_001014450
|
SPRR2A
SPRR2F
|
small proline-rich protein 2A
small proline-rich protein 2F
|
|
chr2_-_228497801
|
9.735
|
NM_001162483
NM_020161
|
C2orf83
|
chromosome 2 open reading frame 83
|
|
chr11_-_62457001
|
9.686
|
|
LRRN4CL
|
LRRN4 C-terminal like
|
|
chr1_-_115323262
|
9.649
|
NM_001102396
NM_025073
|
SIKE1
|
suppressor of IKBKE 1
|
|
chr1_-_201346804
|
9.596
|
NM_000364
NM_001001430
NM_001001431
NM_001001432
|
TNNT2
|
troponin T type 2 (cardiac)
|